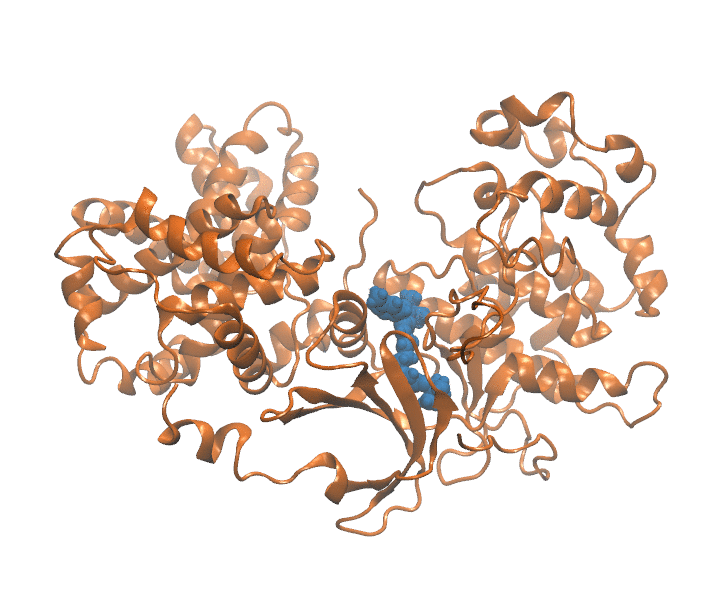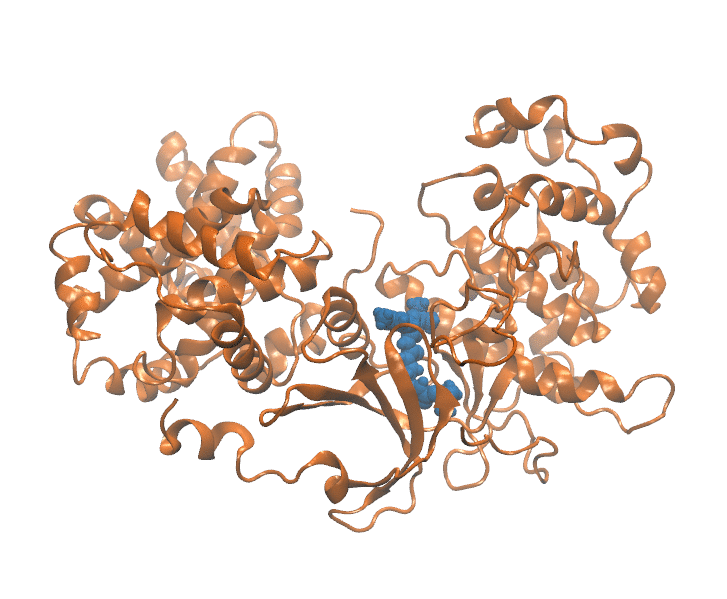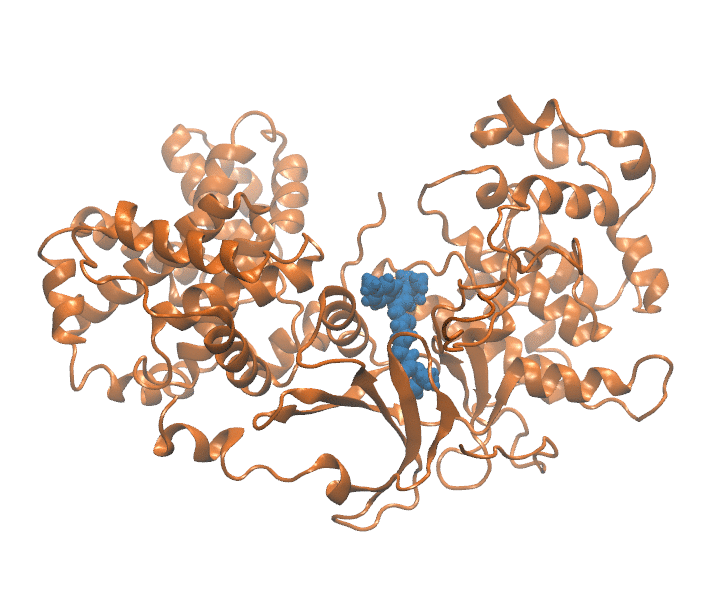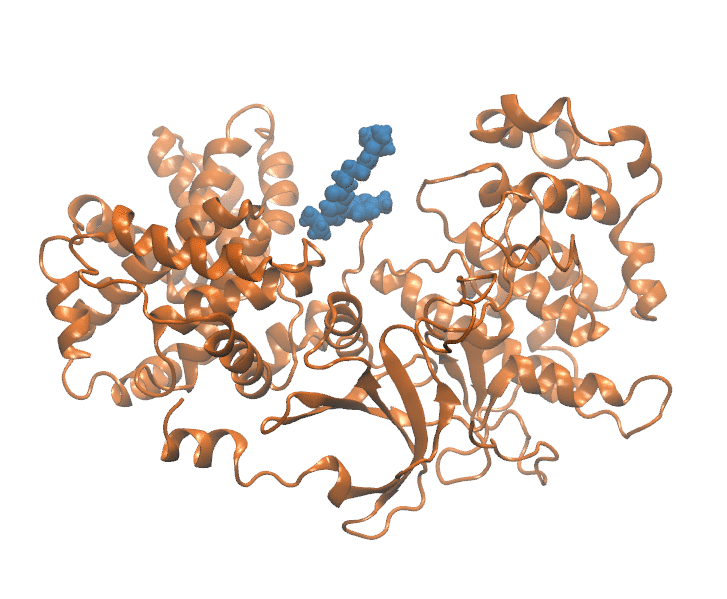
Introduction
Requirements
Downloads
Installation
Introduction
Binding Kinetics Toolkit (BKiT) uses milestoning theory to compute the free energy profile and kinetics properties for a given molecular dynamics trajectory. The toolkit includes newly developed strategies to approximate coordinates for ligand–receptor unbinding or conformational transitions using principal component analysis (PCA). By transforming cartesian coordinates of the complex conformation into a single point in a reduced PC space, we extract major motions essential for describing the kinetics and free energy calculation. Unbinding path is partitioned with disks/lines (milestones) to assign indexes in the PC space. The indexes can be used as milestones or microstates for further investigation.
Requirements
To Run the package
- Python3.6 >
- pytraj 2.0.5 >
Downloads
Installation
-
Clone the repository:
-
git clone https://github.com/chang-group/BKiT.git
-
then run the following command:
-
python setup.py install
References
[1] Tang, Z., Chen, S-H and Chang, C-E. A., Transient States and Barriers from Molecular Simulations and the Milestoning Theory: Kinetics in Ligand−Protein Recognition and Compound Design. J. of Chemical Theory and Computation, 2020, 16, 1882-1895[link]