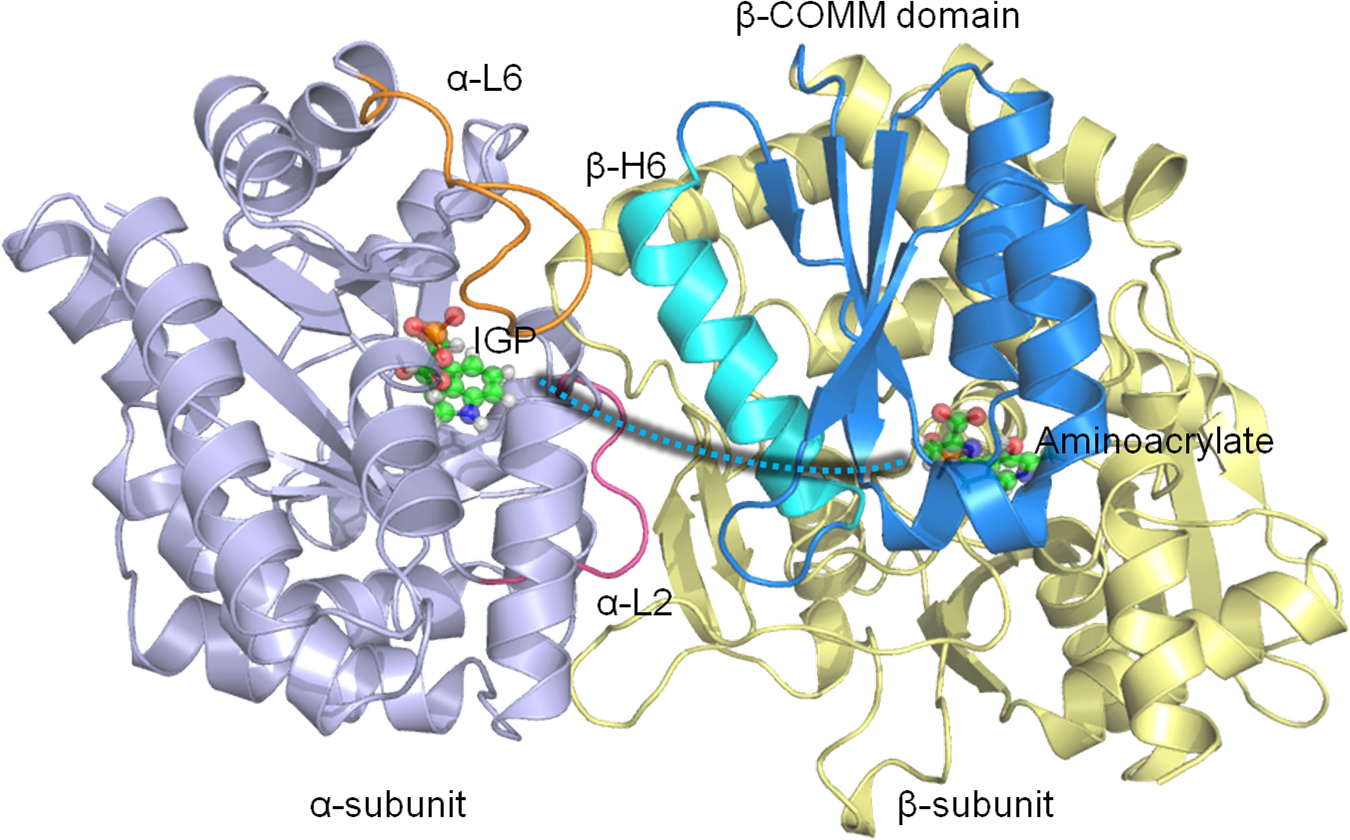
The central goal of our work is to understand the fundamental mechanism of biomolecular recognition and binding kinetics using theory and classical mechanical models. Our research involves the development and application of computational methods and theoretical models to address medically and chemically important problems. These methods are of practical importance in studying biomolecular function, and in the design of new molecules that bind strongly to their receptors. Systems of particular interest include existing or potential drug targets, cell signaling complexes and chemical host-guest systems. Our lab also collaborates with experimental groups on and off campus.

Kinetics of Ligand-Nanostructure Association
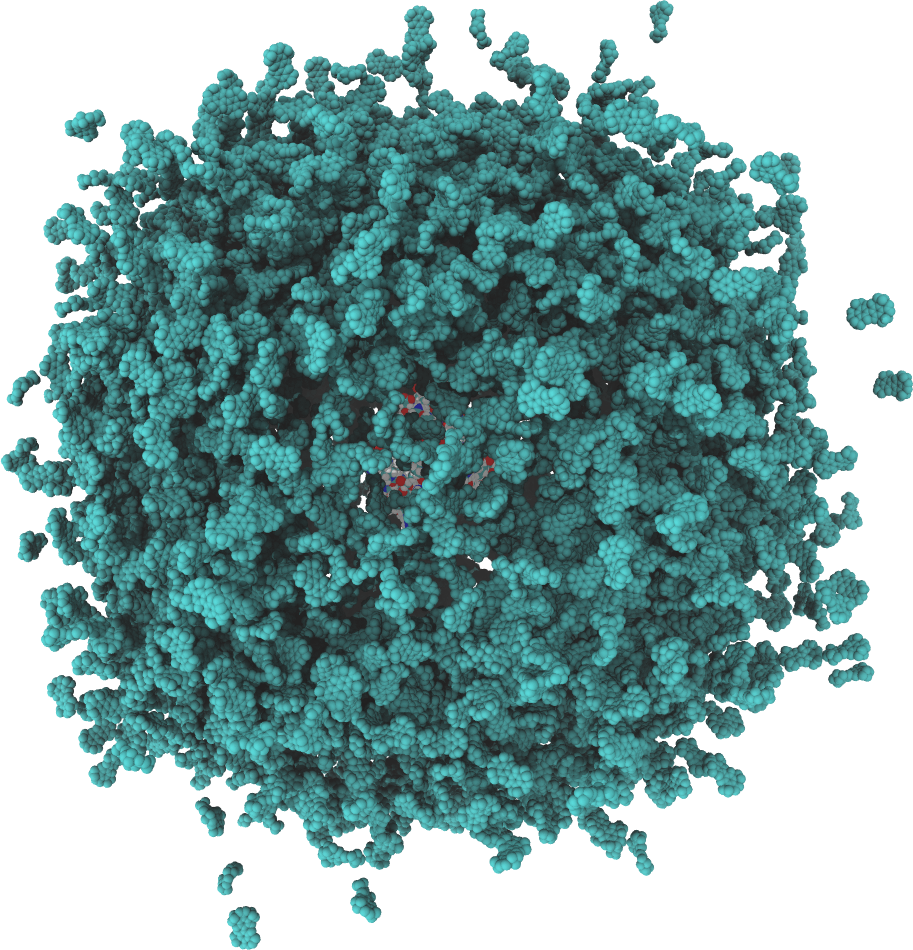
We characterize the kinetics of diffusional enzyme-substrate association. Our software, GeomBD, is designed to assess association rate constants for protein-substrate systems ranging in size from small, single protein systems to large bioengineered multi-enzyme nanostructures.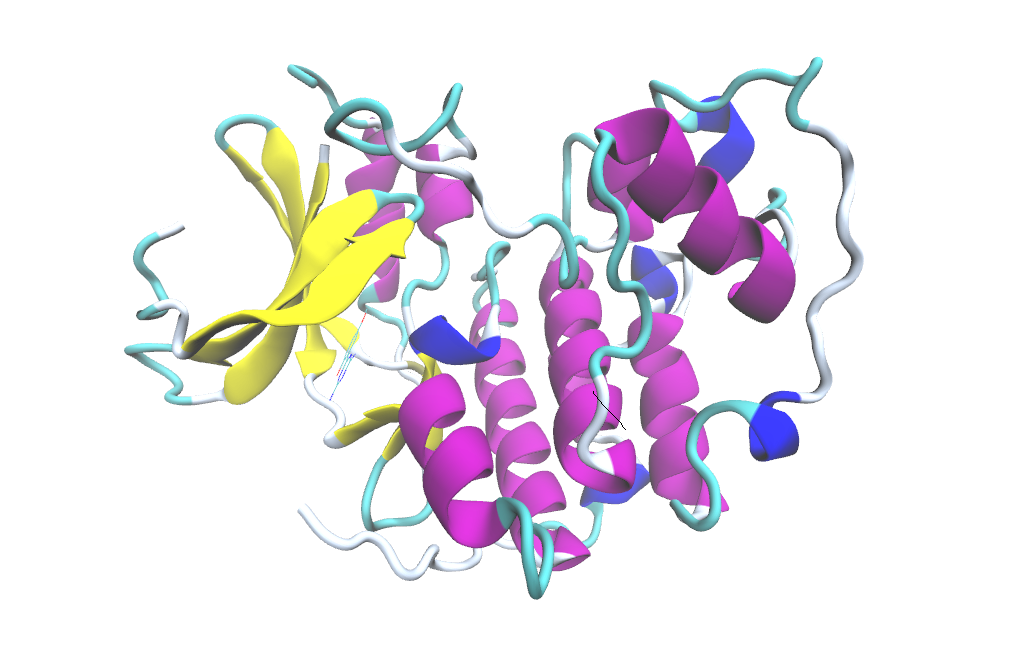
Drug Binding Thermodynamics and Kinetics
The phosphorylated cyclin-dependent kinase 2 (CDK2) regulates cell cycle through the G1 to S phase transition and its deregulation has been implicated in numerous types of cancer. In addition to thermodynamic contributions, kinetic effects are now increasingly considered as important factor in designing efficient inhibitors for targeting CDK2.